What is Bioimaging?
Kim Findlay wanted to be an astronaut staring out beyond our limit of vision into the cosmos, instead she became our Head of Bioimaging, staring down beyond our limit of vision at the microscopic world.
“Bioimaging is a platform technology that is based around microscopy.
Here at the John Innes Centre we offer a mixture of light microscopy which uses light for imaging, and electron microscopy which use beams of electrons instead of light.
Electrons have a much shorter wavelength than light and therefore you can get higher resolution images, although the capabilities overlap, so you could use a scanning electron microscope to image things at the millimetre or micrometre scale or use a light microscope to image things all the way down to a few tens of nanometres. This means you need to choose the best technique to see what you need to see.
Our highest resolving microscope at the John Innes Centre is our Cryo-TEM microscope, which we installed in January 2018. It depends on the sample, but that can resolve down to a fraction of a nanometre.
Because we are imaging biological samples, we are limited by the preparation required. The best preparation we can do is cryo preparation, but this is quite a challenging process, but with an ideal sample and everything working well, we can use the images to determine structures to only a few Angstroms, getting closer to atomic level resolution.
For example, Jake Richardson is currently working with Professor Tony Maxwell on a project looking at the DNA gyrase sub-units of a bacteria, which has been months of work to biochemically prepare and screen the samples by TEM. Jake is using a technique called single particle analysis to ultimately derive the protein structure. After many rounds of experimentation, we have now got some images that are very promising, so these have been passed to Dr David Lawson in the Protein X-ray Crystallography team, who will hopefully be able to use them to determine the protein structure using specialist software.
Ultimately, the goal is to fit an atomic model into the structure and learn how the protein interact with other molecules (e.g. substrates, drugs or DNA), how they undergo structural changes, and how they are structurally-related to other known proteins.
We’re very lucky at the John Innes Centre to have so many centralised services available to help facilitate the science done here, such as protein crystallography or the mathematical processing which is now available, such as that provided by our Informatics team. I’m very proud that microscopy was one of the first core facilities.
Everyone loves putting things under a microscope and we do occasionally get asked to image some weird things. Recently, someone from Professor George Lomonossoff’s group asked us to use our scanning electron microscope to image two clarinet reeds; one used and one unused, after a conversation with someone at an OpenPlant meeting, which inspired a new approach to their work. It is a good example of the creativity and freedom both here at the John Innes Centre and in science generally, to follow the research wherever it might take you.
Clarinet reeds aren’t the only unusual thing we have looked at recently, for example one of our commercial clients asked us to look at some lipstick. As well as covering the John Innes Centre, our microscopes are available for use by anyone, for a fee obviously. We have a lot of users from across the Norwich Research Park, but also lots of commercial clients, from large pharmaceuticals through to North Sea oil companies.
As well as sample preparation, taking the images and training people in how to use the machines, we also use our skills and experience to interpret the images, to help people understand what the images are showing.
For example, when we imaged the clarinet reeds, we were able to clear up some confusion for the scientist around whether some of the images showed two different structures. They were merely seeing two different developmental stages of xylem vessels; which had nothing to do with whether the reed was used or unused, so accurate interpretation of images is vital if you want accurate results.
Additionally, interpreting preparation artefacts can be a problem and even different microscopes can introduce different things to an image, so you need an experienced eye to spot these.
For example, we recently imaged some pea ferritin samples for Dr Janneke Balk’s group, which are particles of about 12 nanometres in diameter; an iron core of about 8 nanometres, surrounded by a protein shell. We put them in the TEM and because they have an iron core, they will scatter electrons, so you don’t need to add any stain, or do any preparation, I just placed it on the grid, dried it and imaged it. You could see a dark iron core, because it scatters electrons so appears dark on the image. I sent Janneke the images and she came back to ask why on some images she could see the protein shell very clearly, but not on others. The reason was merely how I had focussed the microscope, because I know that on the TEM to give you more contrast, you always under focus, which gives you a halo effect and a darker edge, so while it is not such a sharp image, it looks better to the eye because you have that contrast, so you can see the protein shell and the core. However, when at true focus in the microscope, the protein shell disappeared from view, because it was unstained, so all you could see was the iron core. We wouldn’t expect Janneke to know that. This happens all the time; the scientist knows their sample and what they are looking for and then we can help explain what the images are showing, it’s a collaboration.
As a child I wanted to be an astronaut. One of my earliest memories is the moon landing and although I was very young then, it made a big impression on me. The whole world was talking about space exploration and it fascinated me.
From telescopes and deep space, I developed a love of science in general and, in turn, of animals and nature. If not an astronaut I next wanted to be a vet, but as I did my GSCEs and then A Levels, I toyed with the idea of becoming an engineer and going into the Royal Air Force. However, they wouldn’t allow women to sign up for a full commission or go to flying school, so I thought ‘stuff you, I’ll do something a bit different at university’.
I ended up doing a joint degree in biology and physics at King’s College in London and I am so glad I did because it really suits what I am doing now. After university, I almost became a teacher, but I thought I would miss the research aspect of a career in science.
I joined the John Innes Centre as a Research Assistant in the Cell Biology department, which was atypical in that it had its own microscopes. I spent seven years there working on the plant cytoskeletons That was great as it meant I gained a lot of experience in light and electron microscopy. It became obvious that more and more groups would benefit from having a microscopy service they could access, so a case was made for it to become a centralised service available to everyone and I was delighted to be chosen to do just that. Once everyone realised it was available, the floodgates opened and very quickly we needed to expand what we were offering.
It’s developed over time and now we have over 20 microscopes and a team of five and we’re still growing to this day.”
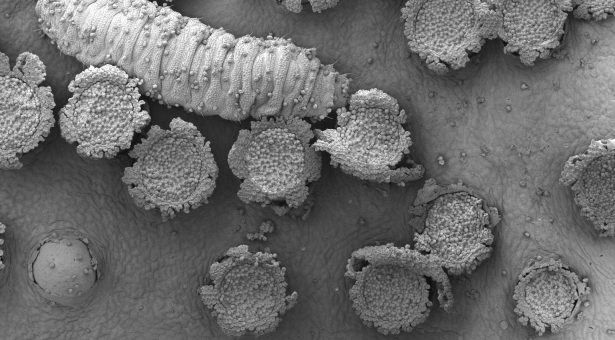
Image details
The image at the top of this page shows rust infection on a Barberry leaf. We are not 100% what the bug eating the spores is, although we believe it is a pollen beetle larvae, if you know different let us know…
