Iron-sulfur cluster assembly in plants
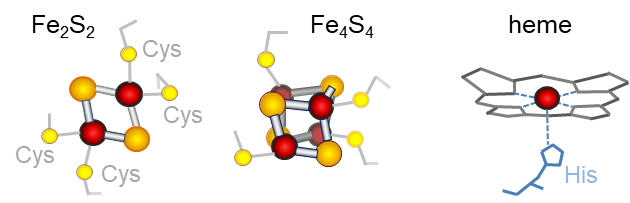
Iron is an important catalyst for many enzyme reactions. The main types of catalytic iron centres (or cofactors) are heme, iron-sulfur (Fe-S) clusters and di-iron or mononuclear iron. In plants, Fe-S clusters are the most abundant (Przybyla-Toscano et al., 2021. Gene atlas of iron-containing proteins in Arabidopsis thaliana, doi: 10.1111/tpj.15154)
The biosynthesis of Fe-S clusters requires specialist assembly proteins that act ‘in situ’, because the clusters are labile and do not exist without a protein fold. However, fully folded Fe-S proteins can be transported across membranes: a nice example is the Rieske protein in plant mitochondria, which requires the Twin-Arginine Translocation (TAT) pathway to cross the inner mitochondrial membrane for incorporation into respiratory complex III (Schäfer et al., 2020; doi: 10.1016/j.cub.2020.01.001).
Plant cells have different pathways for Fe-S cluster assembly in the mitochondria, plastids and the cytosol. The assembly proteins of the ISC (Iron Sulfur Cluster) pathway in the mitochondria and the SUF (Sulfur) proteins in plastids are functional orthologs of those found in eubacteria and cyanobacteria, respectively. However, there are interesting differences, reviewed in Balk & Pilon (2011), Balk & Schaedler (2014) and Przybyla-Toscano et al., (2021) doi: 10.1093/jxb/eraa578.
The Balk group has focussed on the mitochondrial and cytosolic assembly pathways in plants:
- Fe-S cluster assembly proteins in mitochondria
- ATM3, an ABC transporter involved in glutathione export from mitochondria
- Fe-S cluster assembly in the cytosol
Fe-S cluster assembly proteins in mitochondria
The ~12 proteins associated with the mitochondrial ISC pathway are divided in the ‘core assembly machinery’ and downstream carrier proteins.
The core machinery consists of a cysteine desulfurase and the ISU (or ISCU) scaffold protein for the initial assembly of Fe2S2 clusters, also involving a ferredoxin, frataxin and other accessory proteins.
Proteins downstream of the core assembly machinery facilitate the formation of Fe4S4 clusters and transfer the clusters to their final destination. These include GRXS15, ISCAs, NFUs and INDH, which appear to be required for specific Fe-S proteins rather than all mitochondrial Fe-S proteins.
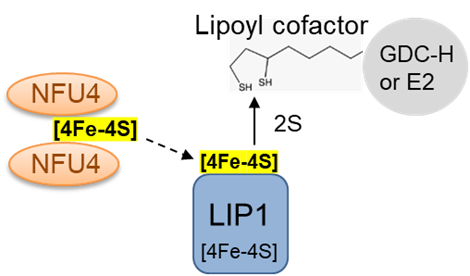
Glutharedoxin GRXS15 is essential for plant viability. However, viable mutant alleles revealed a primary role of GRXS15 in the biosynthesis of lipoate cofactor, which is dependent on lipoyl synthase (LIP1), a rather intricate, Fe-S dependent enzyme (Moseler et al., 2021 and references therein, in particular Ströher et al., 2016 doi: 10.1104/pp.15.01308).
NFU proteins have a highly conserved role in Fe-S cluster assembly in bacteria and eukaryotes. Of the five NFUs in Arabidopsis, NFU4 and NFU5 reside in mitochondria. The two proteins are redundant, but knocking them both out is embryo lethal. A mutant lacking NFU4 and depleted for NFU5 was just about viable, and showed that the proteins have an exclusive role in lipoate biosynthesis (Przybyla-Toscana, Maclean et al., 2021 Plant Physiol), similar to the NFU homologs in bacteria and human mitochondria.
INDH and its homologs in Yarrowia yeast (Ind1) and humans (NUBPL) have a specific role in the assembly of respiratory complex I.
The function of ATM3
After several decades since linking the yeast ‘ABC transporter of mitochondria’ Atm1 and its orthologs in plants (ATM3) and human (ABCB7) to Fe-S cluster assembly, the molecular function of these proteins is still debated.
We showed that yeast Atm1 and Arabidopsis ATM3, when purified from a Lactobacillus expression system, transport glutathione disulfide but not reduced glutathione. The yeast protein was also able to transport glutathione trisulfide, which carries sulfane sulfur (S0), see Schaedler et al., 2014 J Biol Chem).
Arabidopsis atm3 mutants have a more oxidized glutathione pool of the mitochondrial matrix (Schaedler et al., 2014), which we proposed indirectly affects the biosynthesis of molybdenum cofactor precursors (Kruse et al., 2018 Biochem J).
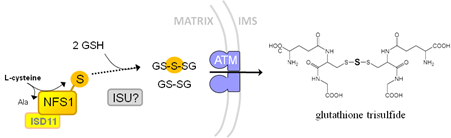
Interestingly, work by Debkumar Pain and coworkers showed that Atm1 exports a sulfur compound and/or Fe-S for tRNA thiolation (Pandey et al., 2018, Cell Chem. Biol; Pandey et al., 2019 J Biol Chem).
Fe-S cluster assembly in the cytosol
The assembly of FeS clusters in the cytosol and nucleus is dependent on the mitochondria. This was first shown in yeast by Lill and coworkers, and the Balk lab confirmed that Arabidopsis NFS1, the mitochondrial cysteine desulfurase, is also required for extra-mitochondrial FeS enzymes (Bernard et al., 2013 Phil Trans Roy Soc B).
The cytosolic FeS assembly pathway (CIA) consists of 8 proteins of which 7 are conserved in plants. All except MET18 are essential for embryo development in Arabidopsis (Bych et al., 2008 J Biol Chem; Bernard et al., 2013 Phil Trans Roy Soc B; Luo et al., 2012 Plant Cell). Viable mutants in NBP35 have serious developmental defects (Bastow et al., 2017 Plant J).
