Regulation of iron uptake by hemerythrin ubiquitin ligases
The iron sensing-signalling mechanism in plants is still poorly understood, in contrast to the well-studied systems in bacteria, yeast and mammals. In particular, we still do not know how the iron status of a cell or group of cells is sensed, but the BRUTUS/HRZ proteins appear to play a key role in this process (reviewed in Rodriguez-Celma et al., Front. Plant Sci. 2019).
The BRUTUS/HRZ proteins form a subfamily of hemerythrin E3 ligases that are conserved from green algae to higher plants. They bear distant homology to FBXL5 in mammals, a post-translational regulator of iron homeostasis. The BRUTUS/HRZ proteins work somewhat differently, in that they target transcription factors for degradation, which in turn activate the expression of genes for iron uptake and mobilization. Thus, BRUTUS/HRZ proteins are negative regulators of iron uptake, and mutants hyper-accumulate iron.
Phylogenetic analysis showed two separate clades in higher plants: the BRUTUS (BTS) and BRUTUS-Like (BTSL) proteins. The rice HRZ proteins belong to the BTS clade. Mutant studies have shown that BTS and HRZ are essential genes, but the BTSL genes in Arabidopsis are not.
The function of BTSL proteins in Arabidopsis
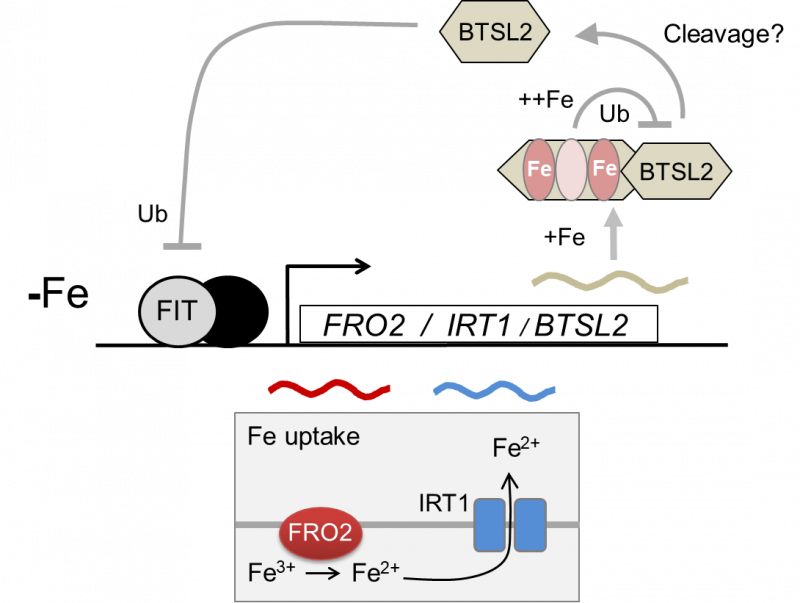
Our in-depth functional analysis of the two redundant BTSL genes in Arabidopsis showed that the transcription factor FIT is a likely substate protein. FIT is a central player in the transcription cascade enabling the response to iron defiency, by inducing the expression of genes for iron uptake. We found that BTSL2 protein interacts directly with FIT in vivo and in vitro, and btsl1 btsl2 double mutants have sustained levels of FIT when moved from iron deficiency to resupply (Rodriguez-Celma et al 2019 PNAS).
We are currently addressing how iron binding to the N-terminal hemerythrin domains of BTS/L proteins affects the E3 ligase activity of the C-terminal domain. This is funded by the Biotechnology and Biological Sciences Research Council (BBSRC), grant number BB/V015095/1, in collaboration with Nick le Brun, University of East Anglia.
The dgl mutation in pea is a small deletion in BRUTUS
The iron-accumulating mutants dgl and brz in pea (Pisum sativum) were isolated several decades ago. Detailed studies of their phenotype have contributed greatly to our understanding of iron homeostasis in plants, but the mutations were never identified.
We used RNA sequencing and exome mapping to link the dgl mutation to a small deletion in a BRUTUS homolog. brz is a point mutation in OPT3, an oligopeptide transporter that mediates iron transport from the xylem to the phloem (Harrington et al., manuscript accepted by Plant J).
Figure: Proposed working model for the regulation of iron uptake by BTSL2