Quantitative regulation of FLC expression

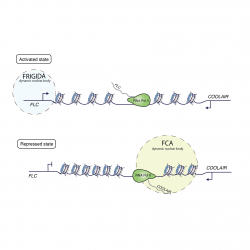
FLOWERING LOCUS C (FLC) expression level is predominantly set by the antagonistic functions of activators and repressors.
FRIGIDA functions with chromatin factors to activate FLC transcription. FCA, 3’ processing factors and chromatin modifiers repress FLC transcription. These repressors influence co-transcriptional processing of FLC antisense transcripts to trigger chromatin modification over the locus and coordinately influence transcriptional initiation and elongation.
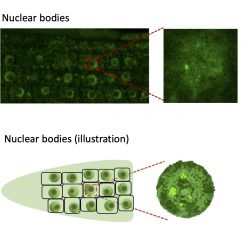
The Dean lab have recently found that both FCA and FRIGIDA form dynamic nuclear bodies essential for different phases of FLC regulation.
Through an EPSRC Physics of Life grant (held jointly with Professor Martin Howard and Mark Leake, University of York) the lab are currently exploring how these phase-separated compartments regulate FLC expression. This is being achieved through a combination of genetic, biophysical and computational approaches.
The lab hypothesise that 3’ RNA processing of antisense transcripts triggers chromatin silencing through an antisense generated R-loop conflicting with the replisome.
They are testing this hypothesis in Wellcome Trust-funded work in collaboration with Dr Julian Sale and Dr Colette Baxter at (MRC Laboratory of Molecular Biology, Cambridge), Dr Yiliang Ding, Professor Crisanto Gutierrez (CMSO, Madrid) and Dr Stefanie Rosa and Anis Meschichi (SLU, Uppsala).