How information beyond the genetic sequence is encoded in the plant sperm
Hereditary information is passed from parent to offspring in the genetic code, DNA, and epigenetically through chemically induced modifications around the DNA.
New research from the John Innes Centre has uncovered a mechanism which adjusts these modifications, altering the way information beyond the genetic code is passed down the generations.
DNA methylation, one example of these epigenetic modifications, happens when a methyl group or chemical cap is added to the DNA, switching a gene, or genes, on or off.
As germline (eggs and sperm) cells develop some of the methyl markers are reset, affecting the information passed onto the next generation.
How this process worked during plant reproduction has until now, been unclear.
The exciting research, published in Science, reveals the molecular mechanism of DNA methylation reprogramming in the male germline of plants.
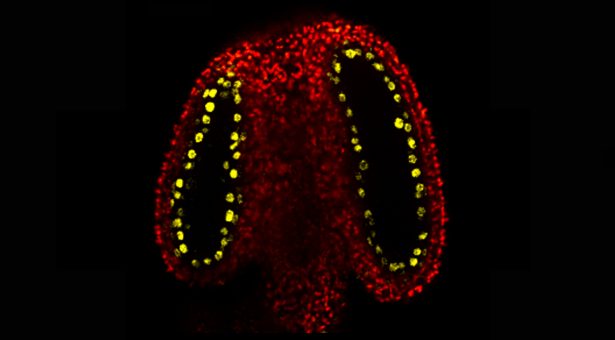
Inside the plant’s male reproductive parts (the anthers), cells that will divide to produce the sperm (meiocytes) are surrounded by cells that nourish them. These nurse cells are called tapetal cells.
The John Innes Centre team discovered that tapetal cells produce an abundance of small RNA molecules and observed that this is caused by a protein called CLSY3, found specifically within tapetal cells in the anther. These small RNAs were shown to move from the tapetal cells into the meiocytes. Here they add new methyl marks to transposons (unstable genetic elements) with the same DNA code.
“This discovery changes the way we think about epigenetic inheritance across generations in plants by showing that small RNAs produced by germline nurse cells can determine the DNA methylome in the sperm. The key role played by these small RNAs in determining the inherited DNA methylome indicates convergent functional evolution between plant and animal reproduction,” says corresponding author Dr Xiaoqi Feng, Group Leader at the John Innes Centre.
This reprogramming stops the transposons from jumping around in the germ cells, and this protects the integrity of the genome between generations.
In the meiocytes, these small RNAs also target genes with similar DNA sequences as the source transposons, helping to control gene expression and facilitate meiosis, a type of cell division that leads to the production of sperm.
The findings have wide application across plant and animal kingdoms and provide a vital new clue for the world-wide community of researchers studying epigenetics.
Previous work has shown that cereal crops, like maize and rice, have similar tapetal small RNAs, however, it was unclear why these small RNAs are important for fertility and yield. The mechanistic insight generated by this study points to new directions of investigations and may help develop biotechnology to target DNA methylation in commercial crops.
Joint first author Dr Jincheng Long said: “Our study could open a new avenue of crop biotechnology. For example, through the manipulation of small RNA directed DNA methylation of the cells that directly contribute to seed formation and the breeding process.”
The study is also important in fundamental biological terms, joint first author Dr James Walker explains, “Our work demonstrates that paternal epigenetic inheritance is determined by tapetal cells, which drive reprogramming at a scale unprecedented in plants.
“The molecular mechanism our work revealed pushes our understanding of de novo DNA methylation to the next level, showing how new methyl marks are established at specific sites in specific cells.”



