How transcription delivers epigenetic silencing
Two new studies which appear in Molecular Cell reveal core principles of how chromatin and transcription interconnect to regulate gene expression.
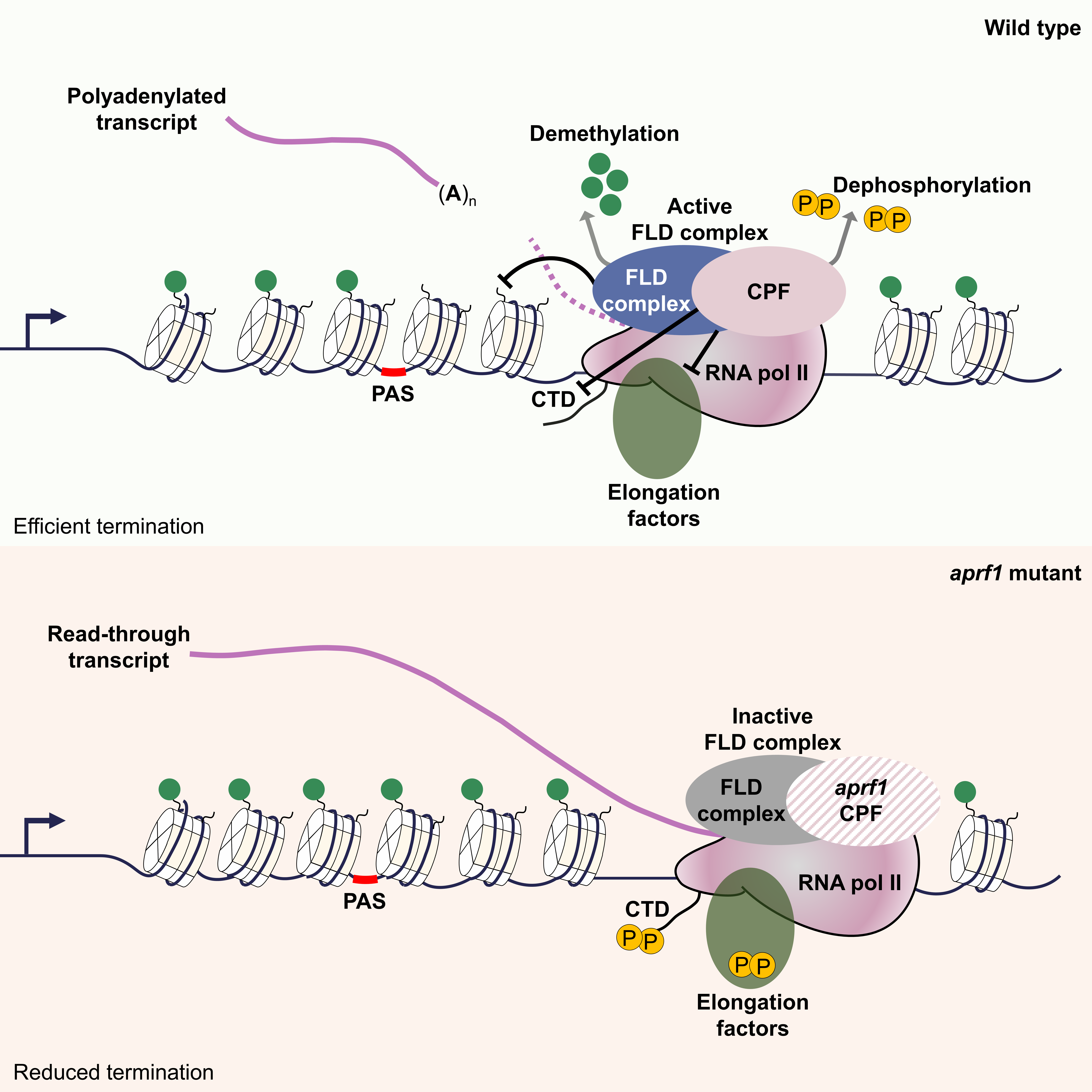
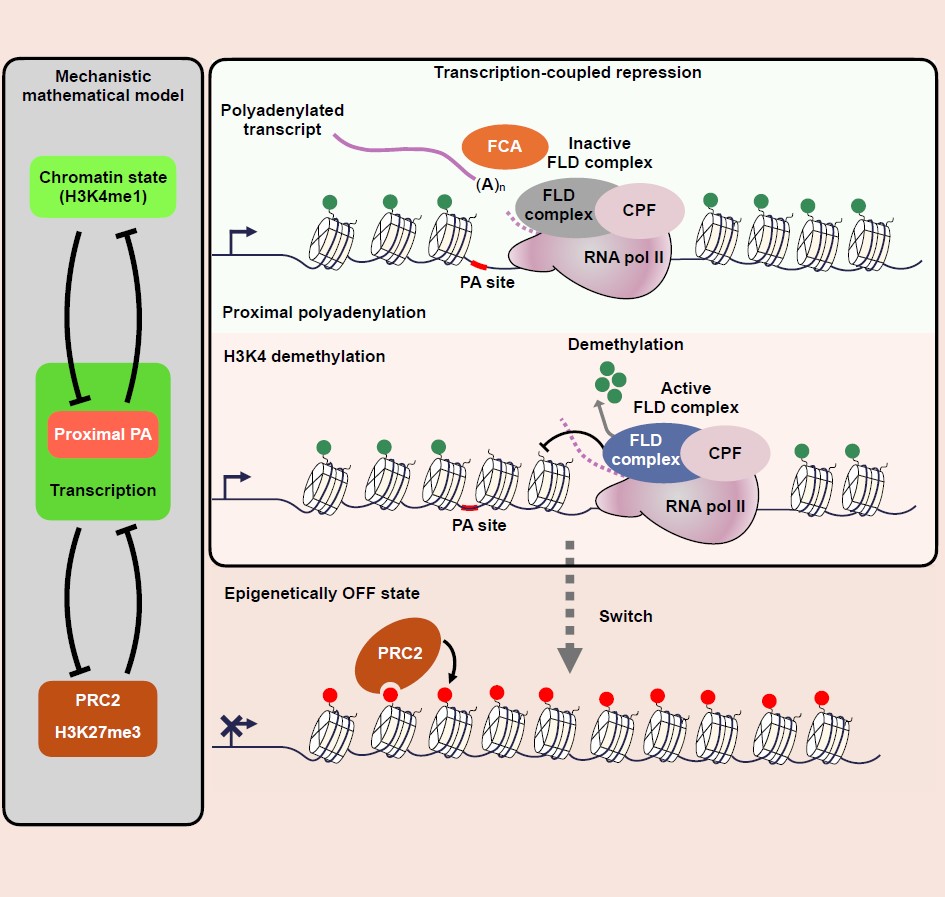
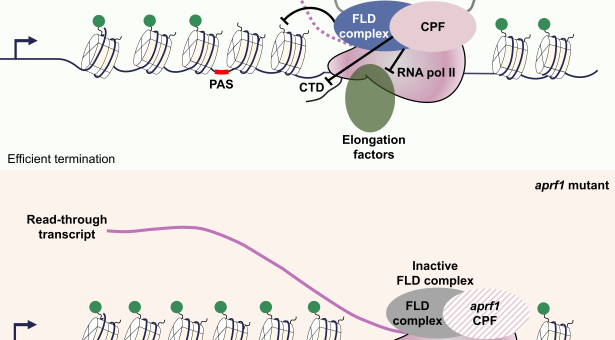
The work was focused on FLC, a gene that determines developmental timing in plants. But the principle that emerges, of the balance between transcription termination and anti-termination regulating gene expression, is relevant to gene regulation in all organisms.
The research by the Caroline Dean and Martin Howard groups at the John Innes Centre, in collaboration with the MRC Laboratory of Molecular Biology (LMB) in Cambridge, shows how feedback between RNA processing, chromatin structure and transcription generate two self-reinforcing modes of regulation – a high or low transcriptional ‘gear’- that can switch from one to another.
It is well known that the transcription of eukaryotic genes can be switched on or off, but how this relates to the structure of the chromatin has been unclear. Many years of research has revealed that the chromatin structure is influenced by the activity of enzymes that deposit and remove specific chemical modifications to the associated histone proteins, but how these influence the switch from ‘on to off’ has been challenging.
Dr Eduardo Mateo Bonmatí, now a Ramón y Cajal fellow at the Universidad Politécnica de Madrid, and lead author of one of the papers explains more, “We uncovered that the transcription of the gene and the processing of the nascent transcript are directly linked to a group of enzymes that modulate chromatin structure.”
“Remarkably, a protein known to regulate FLC for over 20 years, LUMINIDEPENDENS (LD), is central to this process, a fact previously obscured due to its high intrinsic disorder and low amino acid conservation. Thanks to newly developed structural biology prediction tools and our collaborators at the LMB in Cambridge, we discovered that LD is indeed part of a highly conserved machinery present from yeast to humans. While many molecular details remain to be elucidated, our work lays the groundwork for future studies on the relationship between transcription and chromatin.”
While the first paper focusses on how processing of a transcript drives chromatin changes, the second also examines the other side of this interaction, how the state of the chromatin influences the processing of the transcript.
At FLC, these interactions in both directions come together to form a feedback loop. The complexity resulting from having feedback loops has made these mechanisms difficult to unravel until now, even though the core molecular components involved have been known for more than 15 years.
Dr Govind Menon at the John Innes Centre, and lead author of the second paper describes how they unravelled this puzzle, “Mathematical modelling has been the key to dissecting this system. By combining modelling with molecular, genetic, and imaging experiments, and looking over a developmental time course, we were finally able to pick apart these feedback interactions.”
“This allowed us to reveal and quantitatively characterise the molecular mechanisms underpinning FLC switching from on to off by the important Autonomous Flowering Pathway. This study represents the culmination of many years of work on this pathway by the Dean and Howard groups.”
Together these research findings lay out a new and widely relevant paradigm for the interplay between chromatin and transcription across organisms, and how it can shape epigenetic gene silencing.
See a video accompanying these studies, produced by Margot Riggi and Janet Iwasa, University of Utah explaining these molecular mechanisms here – redirects to https://youtu.be/atLsplJ_STk
Links to the studies published in Molecular Cell.
- Mateo-Bonmatí et al. A CPF-like phosphatase module links transcription termination to chromatin silencing. 1016/j.molcel.2024.05.016
- Menon et al. Proximal termination generates a transcriptional state that determines the rate of establishment of Polycomb silencing. 1016/j.molcel.2024.05.014