G-quadruplex Atlas shapes the future for RNA researchers
Dr Yiliang Ding and her group at the John Innes Centre have been awarded prestigious European funding to take forward pioneering research into RNA and its structure. In the second of this two blog series we find out more about this flexible molecule and its role in plant, animal and human health
RNA sequences carry genetic information and fold into complex structures that regulate sophisticated biological functions such as growth and defence.
Among these structures, the RNA G-quadruplex (rG4) is emerging as intriguing and functionally important.
Dr Yiliang Ding’s group at the John Innes Centre were the first to identify the four-stranded RNA rG4 structures in living plant cells.
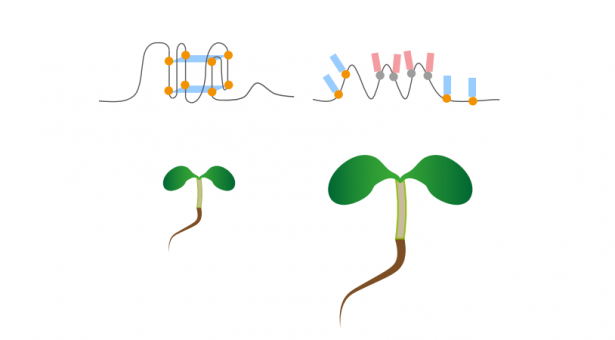
In a recent study, the team carried out genetic and biochemical analyses to reveal that rG4 folding can regulate the fundamental biological process of translation, whereby the genetic code from RNA is translated into protein.
This study which appeared in Genome Biology in September 2020, proposed that rG4 functions in processes which modulate plant growth.
Since then, more exciting discoveries have followed. In recent research which appeared in the journal Nature Communications the Ding group analysed RNA across 1000 plants and observed how recurring structural elements corresponded to habitats.
Interestingly they found that plants growing in cold climates have transcriptomes (the readout of all RNA molecules) enriched in guanine, which are prone to forming rG4 structures.
Immunofluorescence techniques to trace cell activity, supported by experiments on plants, showed that rG4 formation in plants is enhanced in response to cold.
The researchers propose that plants have adopted the rG4 structure to facilitate their adaptation to the cold during evolution.
“Given that we have revealed the presence of a large number of rG4 structures in plants, we are now starting to unravel their individual regulatory roles in plant growth, development and stress response,” explains Dr Ding.
rG4 Atlas
The Ding Group has taken advantage of recent advances to create the first comprehensive transcriptome-wide RNA G-quadruplex database.
G4Atlas is an open-source research tool which will enable a new generation of researchers to find examples of these four-stranded macromolecules in transcriptomes and explain their biological function.
“With recent discoveries on the functional importance of rG4s, a database with experimental validations across diverse species is in great demand. Our G4 Atlas will enable users to explore the general functions of rG4s in diverse biological processes. In addition, G4Atlas lays the foundation for further data-driven deep learning algorithms to examine rG4 structural features,” explains Dr Ding.
The G4Atlas contains data and resources relating to ten species, including bacteria, plants, and animals, 82 experiments and 238 biological samples. Tools include powerful searchable interactive graphs and a wealth of downloadable resources.
Researchers can quickly target their species and/or genes of interest and access detailed rG4 information and resources.
Dr Ding says: “Our work over the past five years has tried to understand the structure of RNA inside the cell. We answered the longstanding question about whether RNA G-quadruplex structures exist in living eukaryotic cells and revealed the presence of many of these structures.
“Now the vast resources of rG4 data in our G4 Atlas is likely to facilitate the emergence of data-driven algorithms in predicting rG4 structure in the future.”
G4Atlas: a comprehensive transcriptome-wide G-quadruplex database appears in Nucleic Acids Research Research.



