History of Streptomyces research at the John Innes Centre
Streptomyces are a group of soil bacteria that are the source of most antibiotics in clinical use today. The John Innes Centre is known as the ‘home of Streptomyces genetics’; here we take a whistle-stop tour through the illustrious history of the research into this bacterium
 1950
1950
Discovery of genetic recombination
David Hopwood (then a PhD student at Cambridge University) was one of the six groups across the globe to discover genetic recombination in Streptomyces– the process of rearrangement of DNA sequences by the breakage and rejoining of chromosomes. He also established a system of genetic mapping in a strain of Streptomyces coelicolor that enabled researchers to identify where genes are located in the genome.
 1961
1961
Early genetic mapping data
David Hopwood moved to the University of Glasgow, and by 1965 his early genetic mapping data had demonstrated a single chromosome in S. coelicolor but left the question of genome topology – linear or circular – unresolved.
 1968
1968
Streptomyces genetics programme launched
Research into Streptomyces genetics at the John Innes Institute began when Hopwood was appointed as Head of Genetics and John Innes Professor of Genetics at the University of East Anglia. When Hopwood founded the Streptomyces group in Norwich, he had just established a detailed circular linkage map of more than 100 Streptomyces coelicolor genes. Streptomyces genetics began to be taken up as a research area by more and more laboratories, including many pharmaceutical companies worldwide.
 1970
1970
Discovering how Streptomyces have sex
Hopwood’s group provided evidence for plasmid-determined mating in S. coelicolor; a key development enabling an efficient natural mating system for genetic analysis.
 1977
1977
Protoplast fusion for improvement in antibiotic production
The John Innes Institute and the pharmaceutical firm Eli Lilly (Indianapolis, USA) independently developed genetic recombination through protoplast fusion in Streptomyces. This discovery opened the way to using protoplast fusion as a very efficient means of combining mutations from different selection lines in antibiotic strain improvement and was used by pharmaceutical companies to great effect.
 1978
1978
The first transformation experiments on Streptomyces
Mervyn Bibb, then a postdoctoral fellow in Hopwood’s group, used the protoplast technology to achieve the first efficient introduction of purified DNA into S. coelicolor, paving the way for the first gene cloning experiments in Streptomyces carried out at Stanford University, California, USA and the John Innes Institute in 1980, revolutionising the field.
 1983
1983
First hybrid antibiotic produced by genetic engineering
Hopwood’s group was the first to isolate a complete set of genes required to make a Streptomyces antibiotic. The genes were then expressed in another Streptomyces host. The John Innes team went on, in collaboration with Japanese and American groups, to produce the first model “hybrid” antibiotic by genetic engineering. Their research had a major impact and in the next decade led to the technique of combinatorial biosynthesis of “unnatural natural products” to make novel pharmaceuticals.
 1985
1985
Streptomyces Manual and Genetic Toolkit
With its success in developing cloning methods, the Streptomyces group actively embraced a training role, offering practical courses based around its influential Genetic Manipulation of Streptomyces: a laboratory manual (now called Practical Streptomyces Genetics). The group went on to develop an extensive toolkit for the genetic manipulation of a wide range of Streptomyces species that has been used widely in academia and industry ever since. Advances associated with this toolkit included the development of a PCR-targeting system for genetic manipulation of Streptomyces (pioneered by Keith Chater and his group) and engineered versions of Streptomyces coelicolor optimised for the heterologous production of specialised metabolites (pioneered by Mervyn Bibb and his group).
 1996
1996
Discovery that Streptomyces chromosomes are linear
Together with collaborators in Taiwan, researchers at the John Innes Centre surprisingly found that unlike most bacteria, Streptomyces chromosomes are linear. This led to an understanding of the novel architecture and replication of Streptomyces chromosomes and had important implications for understanding the role and general occurrence of genome circularity in bacteria.
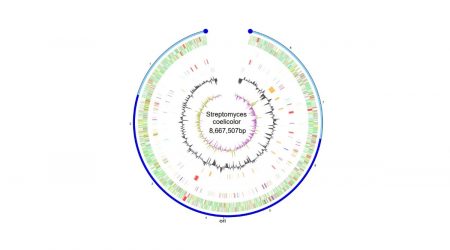 2002
2002
Streptomyces coeilicolor genome sequenced
Scientists at the John Innes Centre oversaw the genome sequencing of Streptomyces coelicolor by the Sanger Centre in Cambridge, and the subsequent development of a range of genomic tools by UK consortia. The sequence demonstrated for the first time that Streptomyces genomes are very large and contain many gene clusters specifying the biosynthesis of unknown but potentially interesting and useful metabolites.
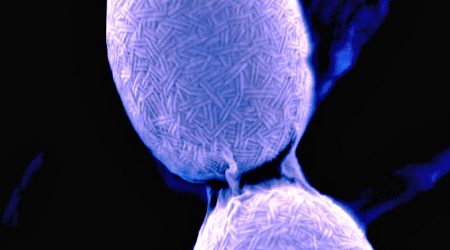 2003
2003
Discovery of the chaplins
Mark Buttner led one of the two groups to discover the chaplins, a family of hydrophobic proteins that coat the aerial hyphae – the specialised reproductive structures that go on to form spores. This hydrophobic coat is required to allow the aerial hyphae to break surface tension and emerge into the air.
 2008
2008
Decoy techniques used to influence gene expression and to take on MRSA
Decoy oligonucleotides were developed and used to influence antibiotic production in Streptomyces coelicolor. These were then applied to counter antibiotic resistance in human pathogens, leading to founding of Procarta Biosystems, a spin out company of the John Innes Centre.
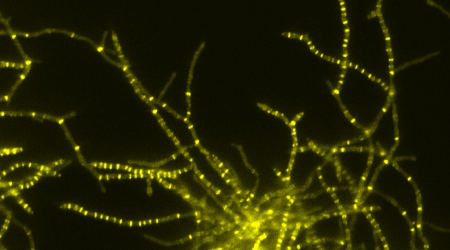 2010
2010
Streptomyces venezuelae as a new model species
The John Innes Centre launched Streptomyces venezuelae as a new model. This species has the unusual ability to sporulate in liquid culture, greatly facilitating the application of global techniques and cell biological methods to the study of development. Streptomyces venezuelae has been widely adopted by labs across the world and is now considered a model strain for the study of morphological differentiation in Streptomyces.
 2011
2011
Novel method for increasing antibiotic yields
Mervyn Bibb and collaborator Koji Yanai from a Japanese laboratory discovered 36 repeating copies of one gene cluster in a strain of Streptomyces kanamyceticus that had been repeatedly selected to over-produce the antibiotic kanamycin. They were able to engineer these components into genetic ‘cassettes’ and then insert these into another Streptomyces strain. They successfully used the cassette system to make Streptomyces coelicolor overproduce actinorhodin, a blue-pigmented antibiotic.
 2014
2014
Sporulation mystery solved
Researchers in Mark Buttner’s lab identified the mystery signalling molecule (3’, 5’-cyclic diguanylic acid) that orchestrates the process of sporulation in Streptomyces bacteria and the unique way in which it works.
 2017
2017
Antibiotic from bacteria found on ant to beat MRSA
Together the groups of Barrie Wilkinson and Matt Hutchings discovered a new structural class of antibiotics made by a new Streptomyces species isolated from an African fungus-growing ant. They named the antibiotics formicamycins and the bacteria Streptomyces formicae after the Latin ‘formica’, meaning ant. They found that these new antibiotics are effective against methicillin-resistant Staphylococcus aureus (MRSA) and Vancomycin-Resistant Enterococci (VRE), bacteria that are resistant to most commonly used antibiotics and that cause life-threatening infections.
 2020
2020
Streptomyces and the smell of soil
It had been known for decades that Streptomycesproduce geosmin, the molecule that gives soil its evocative, earthy smell, especially after rain. Researchers at the John Innes Centre, working closely with collaborators in Sweden, uncovered the role of this compound in nature, showing that it lies in an ancestral mutual relationship between the Streptomyces soil bacteria and primitive, six-legged soil creatures called springtails (Collembola). Geosmin guides springtails to Streptomyces to eat as a privileged food source and in return the springtails help disperse the spores of the bacteria.
