Bacteria can tell the time
Humans have them, so do other animals and plants – now research reveals that non-photosynthetic bacteria too have internal daily clocks that align with the 24-hour cycle of life on Earth.
The research answers a long-standing biological question and could have implications for the timing of drug delivery, biotechnology, and how we develop timely solutions for crop protection.
Biological clocks or circadian rhythms are exquisite internal timing mechanisms that are widespread across nature enabling living organisms to cope with the major changes that occur from day to night, even across seasons.
Existing inside cells, these molecular rhythms use external cues such as daylight and temperature to synchronise biological clocks to their environment. It is why we experience the jarring effects of jet lag as our internal clocks are temporarily mismatched before aligning to the new cycle of light and dark at our travel destination.
A growing body of research in the past two decades has demonstrated the importance of these molecular metronomes to essential processes, for example sleep and cognitive functioning in humans, and water regulation and photosynthesis in plants.
Although bacteria represent 12% biomass of the planet and are important for health, ecology, and industrial biotechnology, little is known of their 24hr biological clocks.
Previous studies have shown that photosynthetic bacteria which require light to make energy have biological clocks.
But free-living non photosynthetic bacteria have remained a mystery in this regard.
In this international study researchers detected free running circadian rhythms in the non-photosynthetic soil bacterium Bacillus subtilis.
The team applied a technique called luciferase reporting, which involves adding an enzyme that produces bioluminescence that allows researchers to visualise how active a gene is inside an organism.
They focused on two genes: firstly, a gene called ytvA which encodes blue light photoreceptor and secondly an enzyme called KinC that is involved in inducing formation of biofilms and spores in the bacterium.
They observed the levels of the genes in constant dark in comparison to cycles of 12 hours of light and 12 hours of dark. They found that the pattern of ytvA levels were adjusted to the light and dark cycle, with levels increasing during the dark and decreasing in the light. A cycle was still observed in constant darkness.
Researchers observed how it took several days for a stable pattern to appear and that the pattern could be reversed if the conditions inverted. These two observations are common features of circadian rhythms and their ability to “entrain” to environmental cues.
They carried out similar experiments using daily temperature changes; for example, increasing the length or strength of the daily cycle, and found the rhythms of ytvA and kinC adjusted in a way consistent with circadian rhythms, and not just simply switching on and off in response to the temperature.
“We’ve found for the first time that non-photosynthetic bacteria can tell the time,” says lead author Professor Martha Merrow, of LMU (Ludwig Maximilians University) Munich. “They adapt their molecular workings to the time of day by reading the cycles in the light or in the temperature environment.”
“In addition to medical and ecological questions we wish to use bacteria as a model system to understand circadian clock mechanisms. The lab tools for this bacterium are outstanding and should allow us to make rapid progress,” she added.
Implications for this research could be used to address such questions as: is the time of day of bacterial exposure important for infection? Can industrial biotechnological processes be optimised by taking the time of day into account? And is the time of day of anti-bacterial treatment important?
“Our study opens doors to investigate circadian rhythms across bacteria. Now that we have established that non-photosynthetic bacteria can tell the time we need to find out the processes in bacteria that cause these rhythms to occur and understand why having a rhythm provides bacteria with an advantage,” says author Dr Antony Dodd from the John Innes Centre.
Professor Ákos Kovács, co-author from the Technical University of Denmark adds that “Bacillus subtilis is used in various applications from laundry detergent production to crop protection, besides recently exploiting as human and animal probiotics, thus engineering a biological clock in this bacterium will culminate in diverse biotechnological areas.”
- The study: ‘A circadian clock in a non-photosynthetic prokaryote‘, appears in Science Advances
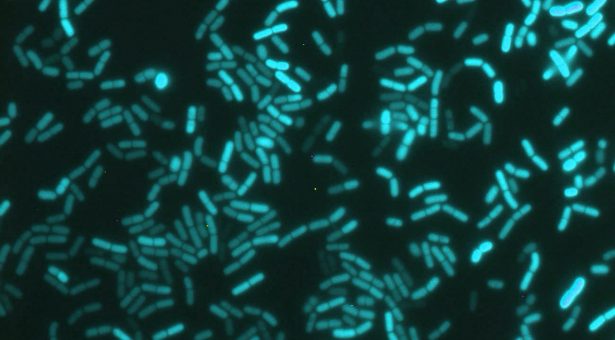
The image at the top of the page was taken by Professor Ákos Kovács (DTU) and is a microscopy image of Bacillus subtilis cells containing a promoter-reporter gene fusion; detected reporter activity false coloured to blue.