How can we use computers to better understand plant development?
We chatted with new Group Leader Dr Richard Smith about his computational path from computer graphics, to investigating how genes control cell growth.
“It is often under appreciated the synergy that exists between computer graphics and scientific simulation.
I started my career in Science in a computer graphics lab that was primarily interested in the simulation of realistic looking plants, In the pursuit of making things look real you often need to think less about describing how a pattern looks, and more about the processes that forms it.
Take the venation pattern in a leaf, for example. It would be pretty hard to specify the dimensions of all the detail of the pattern yourself, but if you can simulate the process of how a leaf venation network is formed mechanistically, then through simulating those processes you can create a realistic looking leaf.
The same is true to make realistic images of clouds, waves in water, or even the wrinkling patterns in Shrek’s clothes as he moves around. Physically based simulation is everywhere in computer graphics, because the better the underlying physics is understood and programmed, the more realistic the animation looks.
Knowledge, methods, and even computer hardware (think GPUs) developed primarily for computer animation can be harnessed to make computer animations of developing plants, which really means understanding the underlying physics of plant development. This is how I ended up a Group Leader in a plant and microbial science research institute despite taking no biology class until I started my PhD aged 41.
Before that, during my undergrad at the University of Regina I met Professor Przemyslaw Prusinkiewicz (Przemek) in a course on digital computer circuits.
This was in the late 1980s, he was running a computer graphics lab and was interested in plants, and most of his lab were working on how to make realistic, life-like computer graphics of plants. A famous example is that of the lilac blossoms they produced in 1988, and I remember being amazed at how real it looked the first time I saw it.
Around the same time, I started a Masters with Przemek, but rather than working on plants, I worked on the development of a real time operating systems for computer music. There were a few musicians in the lab, including Przemek, and we even had a few lab jam sessions, although time for that was scarce. This was before Linux had been invented so I was using the MINIX system as a starting point.
After my Masters I worked in industry for 14 years as an applications programmer and computer consultant. Then at 41, I decided I wanted to do something different, so in 2003 I went back to Przemek’s group at the University of Calgary to do a PhD. By then the group were publishing as much or more in biology journals than they were in computer graphics.
For my PhD I was using computer simulation to model the shoot apex and phyllotaxis, which is how plants arrange leaves and other organs around a stem. You see it in things like pineapples, cactus or pinecones, where you can see these rows of spirals that intersect in different directions. We were interested in how these patterns occurred and what processes were involved in making this happen.
We made models of a growing shoot apex, where genetic networks and cell-to-cell communication determine how plants position organs as they grow, resulting in the intricate spiral patterns that emerge.
We used modeling as a tool for understanding. A biologist might have an idea of how they think something might work; I would like to test that idea in a model. As Information Theorist Gregory Chaitin said; “To me, you understand something only if you can program it.” If a process in development is described in words, then I would like to make a simulation of it, to turn the idea into something well defined, and to really understand it.
I got lucky to jump from PhD to running my own group so quickly. My PhD with Przemek involved a collaboration with Professor Cris Kuhlemeier at the University of Bern, so once I finished, I went over to do a Postdoc with him. I started doing lab work (cloning), but very quickly got caught up in more modelling projects. I’d been there a few months when we got the opportunity to write this grant for a Swiss systems biology initiative called “SystemsX.ch”, in which we proposed a project to look at plant biomechanics.
We were interested in the physical properties of the cells in the shoot apex because if you think about plant growth, ultimately in order to grow, the genes have to change the mechanical properties of the cells. Our focus was less on the genetics, and more about the physical aspects of how the genes implement the biophysical changes required for growth and the emergence of form.
We won the grant, which immediately created a position for me as Assistant Professor, so in a year or so I had gone from being a PhD student to looking after my own group. I never really got to spend much time in the lab, perhaps only long enough to get an appreciation for experimental work. I made a couple of constructs that did get published, but I didn’t get to stay in the lab for long. I liked cloning, it seemed a bit like programming, in that you get to build something.
I worked on the SystemsX.ch Plant Growth project for five years and then at around the time the project was up for renewal, I met Professor Miltos Tsiantis at a workshop. He was very interested in quantifying cellular growth and patterning in Cardamine hirsuta, a close relative of Arabidopsis that has an interesting compound leaf shape. He invited me to join him in his new department at the Max Planck Institute in Cologne.
Ultimately, I spent six happy years there, the Max Planck is a great place to be, but after a while, my family were keen to move to England, so I started to consider what opportunities were available to me here. I felt like the top place in England to continue my work was the John Innes Centre.
In my lab here, we do three things;
- Modelling
- Quantifying the mechanical forces in plants, and
- Quantifying the results of time-lapse confocal imaging
Essentially the latter two are done to support the modelling and we are especially interested in biological processes that involve space and geometry.
Modeling is also often used to address complex genetic networks that involve many components, but restricted to a single cell or compartment, what I call “one-box” models. I become excited when the model contains many boxes (typically cells) with communication between cells. I am interested in how molecular networks and signaling self-organise to create patterns and control their growth to create plant organ shape.
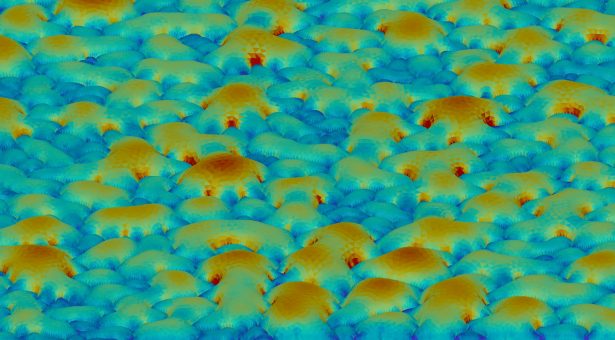
To measure the mechanical properties of plants we developed a device we call the “Cellular Force Microscope” or CFM. It is similar to an Atomic Force Microscope (AFM), a device developed to look at things which are much smaller than the wavelength of light.
The AFM does this by moving a cantilever across the surface and using the forced detected to determine the shape. For this reason it is also known as “scanning probe microscopy”’. We basically made the same thing, but bigger. The name was a bit of a play on words, where AFM seeks to measure things at the atomic scale, we were interested in measurement at the cellular scale. A bit of a joke at first, but somehow the name seemed to stick.
Since plant cells are highly pressurised (double the pressure of a car tire and sometimes more) the force measured with CFM mostly reflects this turgor pressure.
To measure the stiffness of the cell wall, we combine CFM measurements with osmotic treatments to deflate the cells and measure how much they change in size once their turgor pressure has been removed. Cells that shrink a lot are fairly soft, whereas those that shrink less are stiffer.
This is where our imaging work comes in, because in order to measure how much individual cells have shrunk, you need to be able to see them.
Ideally, we would like to image plant organs in full 3D over time. However, this is often not possible in plants, where light scattering limits the depth you can see into the tissue. For flat areas it is sometimes possible to work with a single 2D slice of the image near the surface. However, this is not possible for many organs where there is too much curvature. For example in the shoot apex, the shape of a meristem means that if you just take a single slice of that, you’ll see some cells on the outside of the structure and some on the inside. It doesn’t really give you a picture of how the different cells in different places on the surface are responding.
To tackle this problem, we developed an open-source software called ‘MorphoGraphX’, which takes the sample and extracts the global shape, creating a surface mesh that is used as a curved image. The mesh follows the shape of the meristem onto which the confocal image signal just under the surface is projected.
We like to call these “2.5D” images. They allow you to see and quantify the pattern and shape of the surface cells, and if done over two or more time points you can determine the shape changes of the individual cells on the surface. From this we can calculate cell deformation or growth amounts, directions and cell proliferation.
MorphoGraphX enables the quantification of development in “2.5D” in many organs where it would not possible in 2D or 3D, in both model plants and crop species.
We’re always open to people getting in touch, and to help people quantify development, especially the spatial aspects, such as changing geometry and gene expression patterns.
As fundamental biologists, we aim to understand how cells control their growth and patterning, and how genes control the biophysical properties of cells in order to create the emergent form of an organism.”



