Spatiotemporal protein dynamics
The Howard group has been a pioneer in the application of mathematical models to spatiotemporal protein dynamics.
Past work has included the first model of the MinCDE oscillations regulating cell division positioning in E. coli (Howard et al, 2001), the first model for the dynamics of the Par proteins, and their interaction with the cytoskeleton, in the one cell C. elegans embryo (Tostevin and Howard, 2008), and the first model for the dynamics of hyphal branching in Streptomyces (Richards et al, 2012).
This research has clearly demonstrated that intricate patterns and localisation can be generated from very simple protein interactions and diffusion.
The Howard group has also pioneered the study of low copy number fluctuations in spatiotemporal protein dynamics, as applied to both the MinCDE system (Howard and Rutenberg, 2003) and also to models of morphogen gradient formation, both developmental (Saunders et al, 2012) and intracellular (Tostevin, ten Wolde, Howard, 2007).
The group has also investigated the spatiotemporal dynamics of the ParABC system which regulates low copy number plasmid segregation in E. coli. This is the most common system that mediates plasmid DNA inheritance in bacteria and compromises the proteins ParA and ParB, and DNA sequence parC. Remarkably, these components can position plasmids at equally spaced positions throughout a cell to ensure plasmids are present in both daughter cells when the cell divides into two.
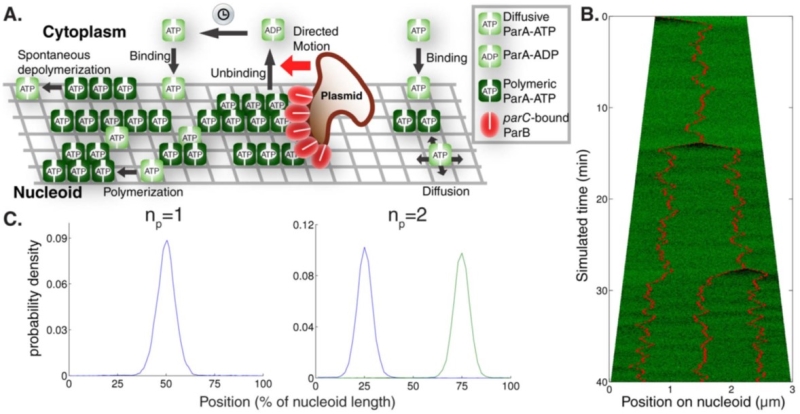
The mechanism by which ParA structures achieve this precise positioning was studied using mathematical modelling and precise experimental measurements.
The group showed that ParA can direct relatively immobile plasmids over the bacterial chromosome using self-organising, competitive ParA structures, whose disassembly is induced by plasmid parC-bound ParB. More generally these findings will help the group understand transport and regular positioning of intracellular cargo.
